CLL transcriptome data
For all considered CLL datasets (Table S3, refs. 38,39,40), we first performed gene expression filtering using statistical distribution fitting and threshold-based filtering (Fig. S1, Methods)14,41. From the whole…
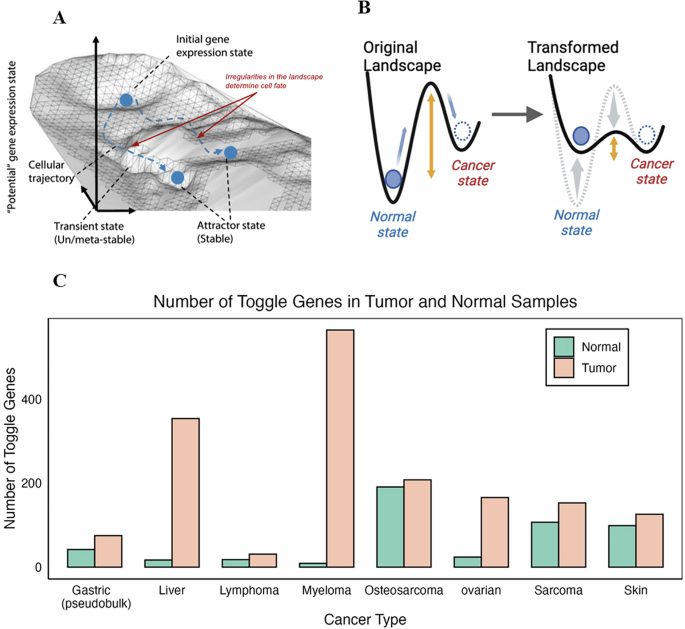
For all considered CLL datasets (Table S3, refs. 38,39,40), we first performed gene expression filtering using statistical distribution fitting and threshold-based filtering (Fig. S1, Methods)14,41. From the whole…
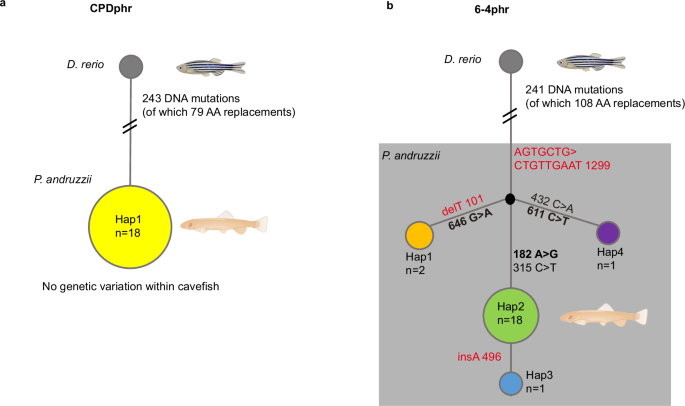
Wild type (iCab) and photolyase mutant medaka (Oryzias latipes) were maintained in the fish facility of the Institute of Biological and Chemical Systems, Biological Information Processing (IBCS-BIP) at…
The murine RAW264.7 cell line was obtained from the American Type Culture Collection (ATCC #TIB-71). RAW264.7 cells were cultured as described elsewhere14 in complete DMEM (containing 1% v/v P/S, 10% v/v FBS, and 1% v/v glutamine) in…
Human eggs can remain viable for decades by dialing down their internal recycling systems, new research shows. Studying over 100 freshly donated eggs, scientists discovered that slowing protein breakdown helps minimize harmful byproducts that…

Three rescued groundhog (Marmota monax) siblings were joyfully released back…
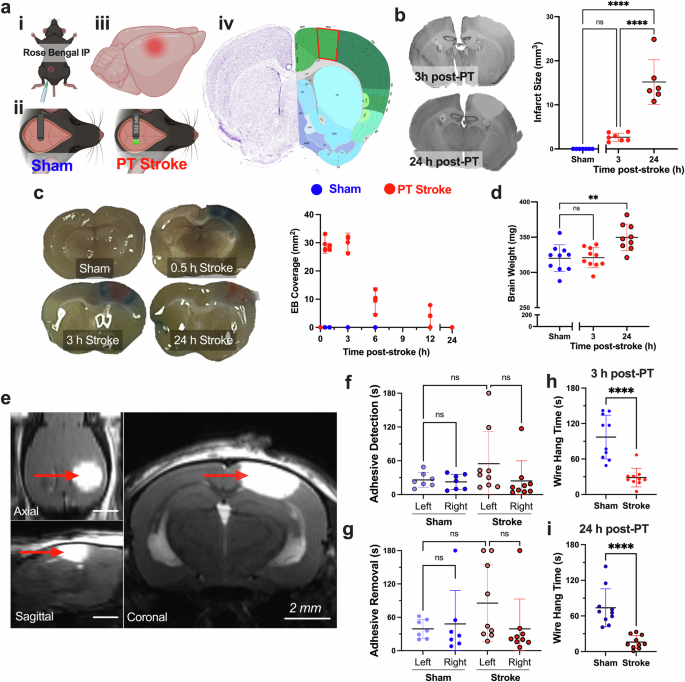
DiSabato, D. J., Quan, N. & Godbout, J. P. Neuroinflammation: the devil is in the details. J. Neurochem 139, 136–153 (2016).
Google Scholar
Gauberti, M. et al….
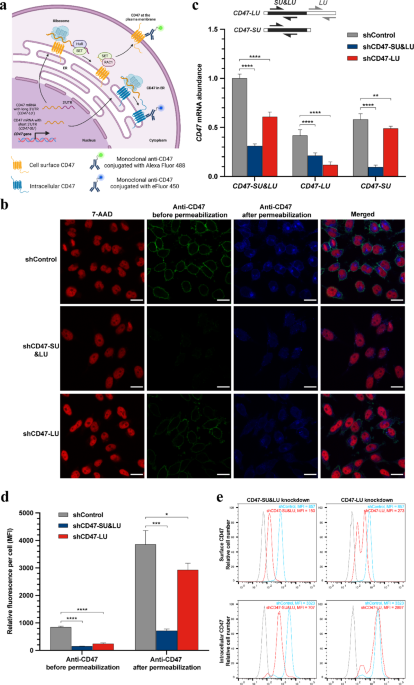
Previous studies have demonstrated that global alterations in mRNA polyadenylation, resulting from the utilization of different PASs, play a crucial role in post-transcriptional gene…
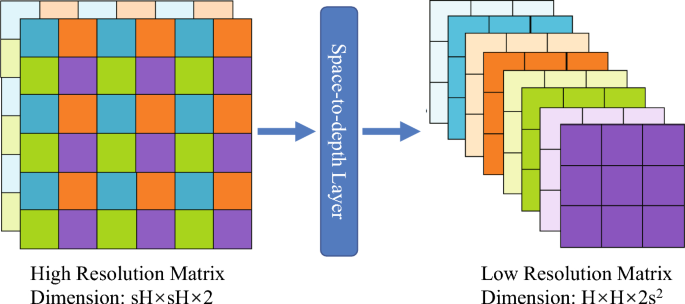
This section focuses on the method of the proposed model. We first introduced the class-specific residual attention module and then proposed a multi-attention mechanism to improve the ability of small object detection (lesions detection). Then,…
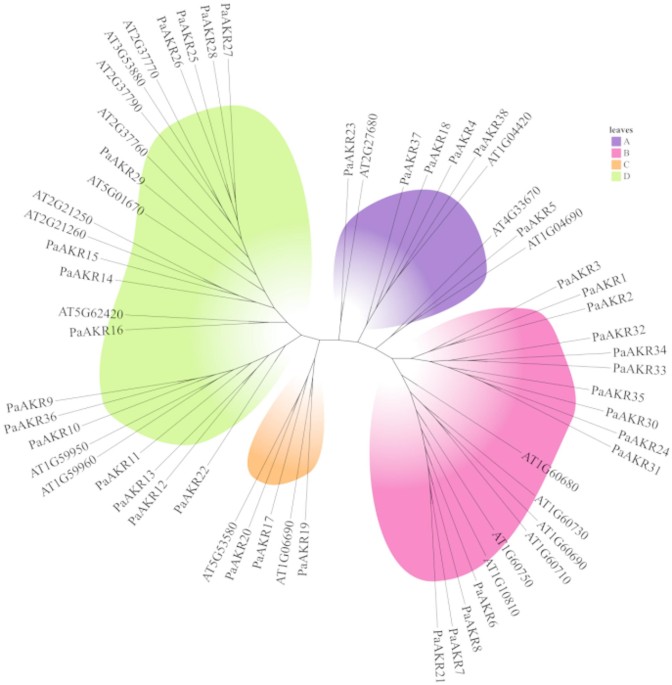
To explore the evolutionary relationships of PaAKR gene family, a phylogenetic tree of 60 genes from cherry and Arabidopsis was constructed using MEGA7.0, which was divided into 4 subfamilies…
Vogel, C., Bashton, M., Kerrison, N. D., Chothia, C. & Teichmann, S. A. Structure, function and evolution of multidomain proteins. Curr. Opin. Struct. Biol. 14, 208–16 (2004).
Google Scholar